Introduction
The following guide is intended to help you think about the various types of evidence you should consider as you attempt to annotate genes in Drosophila virilis, Drosophila mojavensis, or Drosophila grimshawi. The same considerations will also apply to species that are more closely-related to Drosophila melanogaster. However, in many of these species the level of conservation will be high enough that some of the other forms of evidence will rarely need to be considered. The “Annotation Strategy Guide” available on the GEP website is the sister document to this one, and it walks through several annotation examples demonstrating the use of many of the considerations discussed below.
Please note that the last page of this document is a summary sheet for quick reference.
A. Information for Beginning Annotators
Annotation
In practice, creating a gene model is simply designating a series of base coordinates that describe the structure of the gene (i.e., the exact base where each exon begins and ends). In species where evidence of expression (RNA-Seq data or cDNA sequences) is available, one may be able to identify the coordinates of the full-length transcript including the 5' and 3' untranslated regions (UTRs). Promoter and terminator sites are hard to identify so, in projects without data on expression, it is usually very difficult or impossible to annotate the 5' and 3' untranslated regions. As such, for projects without expression data, only the CDS’s (i.e., the regions of each exon that actually code for protein) are described in the gene model.
As an annotator, your job will be to create a gene model that uses as much evidence as you can gather and synthesize that evidence in a thoughtful and biologically consistent way. Your goal is to create a gene model that is consistent with what is known about basic biology and is best supported by the evidence at hand. You probably already know quite a bit of biology that will help you annotate. For example, since it is known that RNA polymerase does not hop back and forth between the two strands of a double stranded DNA molecule, you know your gene model should not include sequences from both strands unless there is experimental evidence that the gene undergoes trans-splicing (e.g., mod(mdg4) and lola; see McManus CJ et al. 2010. PNAS 107:29). In most cases, the gene must start on one strand, continue down the length of that strand and end on the same strand, demarcating the base position of each CDS. For species sufficiently close to D. melanogaster (e.g., Drosophila erecta) or species with sufficient expression data, it might also be possible to identify the non-coding parts of the mature mRNA. However, the principal goal of the GEP annotation projects is to carefully identify the coding regions in each gene, so the discussion below will focus on that goal.
Your first step in annotation will be to collect and consider all the information or evidence you can about the sequence you are annotating. Most, but not all, of the useful evidence has been gathered together for you in the GEP UCSC Genome Browser. When annotating you will probably also want to collect other evidence yourself. Once you have gathered the available evidence, each piece of evidence should be weighed against all the other evidence and used to make your gene model. The goal is to make the best gene model you can that integrates all the collected evidence in a way that maximizes the use of high-quality evidence, avoids internal conflicts, and only uses low quality evidence when no higher quality evidence can be found.
The types of evidence used fall into three basic categories: expression, conservation, and computation.
Expression evidence is derived from the sequencing of RNA that has been isolated from the organism of choice (or an extremely close neighbor species). The sequences are typically mapped back to the genome to provide evidence of transcription. When no expression evidence is available, conservation will be your most important evidence in constructing a gene model.
Conservation evidence relies on the assumption that the new species being annotated has a recent common ancestor with D. melanogaster. Based on the principle of Occam’s razor, which declares that the best explanation for anything is the one with the fewest assumptions, the best gene model in a new species would be the model that assumes the fewest differences (i.e., mutations) between it and the gene model in the very carefully annotated D. melanogaster.
The third general type of evidence is computational. Many computer programs have been written that attempt to recognize various features in DNA. These programs have already been run on the sequences you will be annotating and the results are available for viewing on various genome browsers. These programs are designed to identify evidence for a wide variety of biological features including genes, repeats, or various other features (e.g., intron/exon boundaries). Each of these programs has been worked on and optimized for its given purpose and as such, provides at least a hint as to a possible biological function of any given sequence. Without expression and conservation evidence, computational analysis is usually the only evidence you can fall back on to create your gene models.
Finally, if expression, conservation, and computational evidence fail to provide enough evidence for a given gene model, a few simple rules can be used to assist in creating a gene model. These rules are based on philosophical consideration of how best to “get things wrong” and are discussed at the end.
Basic Biology
Before we consider types of evidence in more detail, we will discuss a few details of basic biology that will guide you in your generation of a gene model. While it is impossible in a short discussion here to cover all the relevant basic biology (you should already know about transcription and translation), there are a few specific details that should be discussed.
Introns. Unlike bacteria, many genes in eukaryotes have introns. These sequences are removed from the primary RNA transcript based on sequences found within the intron. The sequence at the beginning, or 5' end of the intron, is called the donor site, while the 3' end is called the acceptor site (see Figure 1). The consensus sequence that defines donor and acceptor sites has a lot of tolerance for mismatches and can evolve quite quickly.
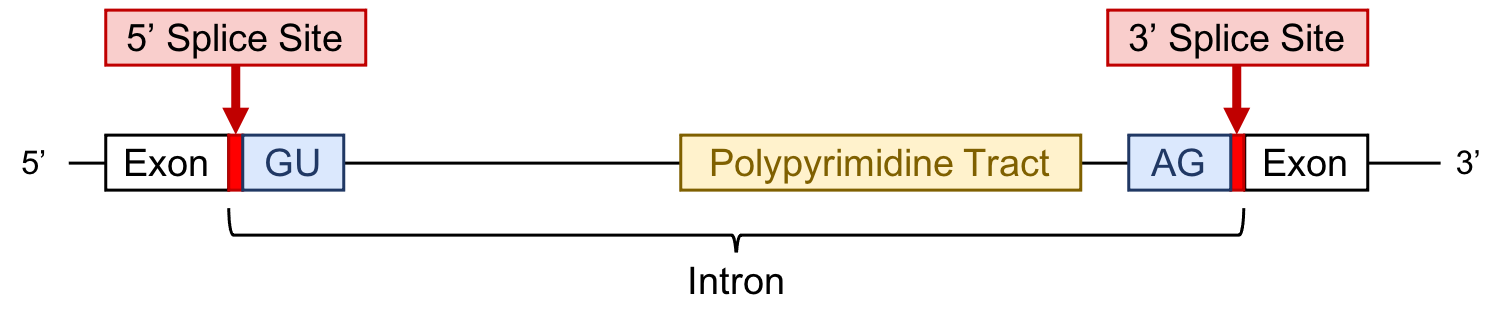
For the purposes of your analysis you can identify putative intron/exon boundaries in three ways. First, you can assume that any de novo gene prediction program will predict donor and acceptor sites consistent with the basic biology of splice site sequence composition; thus, any gene model generated by a de novo gene prediction program can be used to help you pick splice sites. Second, a computer program designed to find and score putative donor and acceptor sites has been run on all sequences. The results of this analysis are displayed in the GEP UCSC Genome Browser for your sequence in the “Predicted Splice Sites” track. To simplify the results, the potential sites predicted using this program have been split into high, medium, and low qualities; sites of any quality can be used as part of your gene model. Finally and probably the least reliable way to find donor and acceptor sites is to look at the sequence by eye and scan for the base sequences known to be used by the splicing mechanism. While searching by eye is the lowest quality of evidence for the prediction of an intron/exon boundary, it is sometimes the only evidence for a given splice site when expression and conservation evidence are lacking. It is important to note that when looking at the genomic DNA (i.e., in the GEP UCSC Genome Browser) the splice donor site sequence will be GT, and not GU as depicted in the above figure of the precursor mRNA. Any GT can be considered as a potential intron donor site, while any AG is a possible acceptor site.
In rare cases (in the range of about 1%), a “non-canonical” donor site with the sequence GC has been detected in D. melanogaster. Non-canonical donor sites will never be used in gene models generated by the de novo gene predictors; however, evidence collected so far in annotation of D. virilis suggest that these GC donor sites are used in a few genes in this species. Of the non-canonical sites found in D. virilis, about 50% of the time the same non-canonical site is used in D. melanogaster. Other non-canonical sequences have been described in the literature, but they are extremely rare (see Table 3 in Crosby MA et al. 2015. G3 5:8). Hence an annotator would need evidence from well-controlled wet bench experiments (e.g., splice junction predictions derived from multiple spliced RNA-Seq reads) before using them in a gene model.
Finally, past studies of introns in D. melanogaster put the minimal size of an intron at 43 bases (see Guo M et al. 1993. Mol Cell Biol 13:1104 and Talerico M and Berget SM. 1994. Mol Cell Biol 14:3434). This intron size limit is supported by the FlyBase D. melanogaster annotation release 6.66, which shows that 99.97% (72041/72062) of the introns are at least 43 bases. It is reasonable to assume that this limit also exists in the other Drosophila species. Thus, any gene model that predicts the presence of an intron smaller than 42 bases is very likely incorrect and a different gene model which does not have such a small intron should be made. In addition, a recent study shows that approximately half of the D. melanogaster introns are between 40–80 bases in length, and that introns in the range of 60–70 bases (the most common intron size range in D. melanogaster) show the highest splicing rate (Pai AA et al. 2017. eLife 6:e32537).
mRNA structure. Once you have the location of the start codon, the stop codon, and all of the intron/exon boundaries, it is possible to predict the final coding sequence of the mRNA as well as the predicted amino acid sequence of the encoded protein. Remember that in eukaryotes each mRNA contains a single open reading frame (ORF) that extends from the start codon through all the internal exons and ends with a stop codon. Your gene model should likewise produce a putative message that contains a single long open reading frame with no internal stop codons. If your gene model has internal stop codons, you should double check and adjust your intron/exon boundaries until no internal stop codons are found.
Expression
Evidence of expression from cDNA/EST/RNA-Seq tracks can provide strong evidence for the locations of intron/exon boundaries. However, there are important things to consider when evaluating expression data. It is important to remember that some genes are expressed at very low levels or only for very short periods of time. As such, the lack of expression data for any gene should not be considered strong evidence discounting the presence of a gene. No matter how much expression data was collected, there will always be real genes that lack expression evidence. The other issue to be aware of is the high level of noise found in RNA-Seq data. We have observed in RNA-Seq data that has been mapped across an intron (i.e., predicted splice junction) a non-trivial amount of variation in the apparent splice site. In situations like this, where there is a lot of variation in the evidence for the exact position of a splice donor/acceptor site, a “majority rules” approach is usually prudent.
Conservation
Conservation can take many forms, and all of these should be considered when generating your gene model. They are presented here in order of importance with the most important first:
Conservation of primary amino acid sequence. This is certainly the most important form of evidence that will guide you in construction of your gene models. It is reasonable to assume that for almost all genes found in the various Drosophila species, the encoded proteins are serving some kind of function and are under selective restraint. As such, one would expect the amino acid sequences of functional proteins to evolve more slowly than sequences that are not functional. This slower evolution leads to the expectation that functional sequences will, in general, show higher levels of similarity. Conservation of this type is found using computer programs like BLAST. When conservation between the two species is very high, the identification of intron/exon boundaries can be easy, as these boundaries will be very close to the end of the alignment (assuming that you are searching with single exon sequences). As the extent of amino acid conservation goes down (in more distant species like D. virilis), the identification of intron/exon boundaries will need to rely on other evidence as discussed below.
B. Information for Intermediate Annotators
Searching for conservation of sequences when the default BLAST search fails. While blastx or tblastn are your primary tools for searching for conservation, just like all other programs, they have their limits and can fail. This will often happen when you search with smaller exons or with sequences that are evolving rapidly. If you do a search and no significant similarity is found, your first step should be to increase the Expect threshold. Remember you are looking for “the most similar” sequences not necessarily “a statistically significant” match. The technique then is to keep stepping up the Expect threshold until you get alignments, no matter how large the absolute value of the E-value. As you increase the Expect threshold, you are allowing for weaker and weaker evidence to be listed in the results; however, weak or very weak evidence is better than no evidence at all so an exhaustive search is best. Any alignments in the proper location (between known exons and on the proper strand) should then be checked to see if the evidence they provide can help you create a viable gene model.
If BLAST fails (and it will in at least some cases), the next best search technique is to use the EMBOSS Water tool to do a DNA-to-DNA search. This is a very different kind of computational algorithm that can sometimes succeed when BLAST fails. The recommended technique is to extract the DNA sequence of only that region of the contig that you wish to search (you must use the proper strand in a Water alignment) and compare it to the DNA sequence of the exon you are trying to place. The result will always be a single best alignment as determined by the Smith-Waterman algorithm. The position of this alignment should then be checked to see if there is sufficient evidence for an exon (i.e., viable donor/acceptor sites).
If the EMBOSS Water search fails, then DNA-to-DNA blastn should be attempted. To avoid large numbers of irrelevant and misplaced alignments, be sure to either extract the region to be searched or use the “Subject subrange” feature at NCBI BLAST. Here again, the Expect threshold should be increased in increments until alignments are found. The alignments generated are again good starting points for further investigation.
Evidence for conservation can also be found in the multi-species conservations tracks currently found in the “Comparative Genomics” section. The “Drosophila Conservation” and “Drosophila Chain/Net” tracks rely on different programs that look for sequence conservation by comparing multiple species. These multi-species examinations can often be more sensitive than a typical BLAST search.
Conservation of gene structure. The creation or removal of an intron in orthologous genes is a very rare event, even over evolutionary time scales represented by these Drosophila species. This means that the best gene model in the target species (e.g., Drosophila biarmipes) will almost always have the same basic structure (i.e., number of introns and exons) as the gene model in D. melanogaster. This rule however is not absolute; sometimes the only gene model that fits most of the evidence has a new or missing intron, so if you can find no way to construct a gene model that maintains the number of exons, go with a gene model that keeps the total number of exons as close to D. melanogaster as possible.
Conservation of exon length. In a surprising number of cases, we have found exons that have a very similar length even when there is no detectable conservation of the encoded amino acids found near the intron/exon boundary. This has happened enough that we can come to consider more carefully any putative donor or acceptor sites that conserve exon length. For example, consider the following alignment in which blastx was used to find similarity between a piece of D. virilis genomic DNA sequence and the sequence of a 45 amino acid long exon from D. melanogaster:
Exon sequence:
1 CGSVVPSADYAYSPAYTQYGGTYGSYSYGTSSGLIYNPAS
41 GPITT
BLAST alignment:
Query 14253 CGSVVPSADYAYSPAYTQYGGTYGSYSYGTSSGLI 14357
C SVVP +DYAY+PAYTQYGG YGSY YGT SGLI
Sbjct 1 CSSVVPGSDYAYNPAYTQYGGAYGSYGYGTGSGLI 35
In this case, we can see that the alignment starts out well (amino acid 1 of the exon is aligning to base 14253 in the sequence being annotated). However, the alignment ends at amino acid 35, well before the end of the exon at amino acid 45. The alignment is missing the last 10 amino acids (remember that BLAST only gives a local alignment; that is, it does not report sequences that do not have significant similarity). In cases like this, we would concentrate our search for donor sites around base 14387 (30 bases or 10 codons down from the end of the alignment). While any donor sites downstream of 14357 (the end of the above alignment) would be a potential candidate, donor sites found near 14387 would be strong candidates for use in the final gene model, especially if they are supported by other observations.
Computational evidence
While expression and conservation will, in most cases, be the best evidence for constructing your gene model, you may run across cases in which expression and/or conservation will give support for several different gene models with no way to pick among the consistent models. In these cases, computational evidence is your next best source. The recommended approach is to rely on expression and conservation as much as possible and adjust your models based on the computational evidence as needed. There are two main sources for information you will want to consider as you try and determine the best gene model, splice site prediction programs and ab initio gene finders.
Splice site prediction program. The “Predicted Splice Sites” tracks on the GEP UCSC Genome Browser shows the results of the splice site prediction program GeneSplicer. The output of this program tags potential splice donor and acceptor sites and gives each potential site a score between -10 and +10. In order to simplify the output, we have classified those sites with scores above 7 as “high quality,” scores between 0 and 7 as “medium quality,” and scores between -10 and 0 as “low quality.” In general, this information can be used to help you pick donor/acceptor sites when there is no expression or conservation. For the purposes of the GEP project, you should always pick a donor/acceptor site that maintains the open reading frame and maximizes conserved amino acids. However, when there is little or no conservation or there are two or more possible donor/acceptor sites very close together, sites which have been tagged by GeneSplicer are better choices than sites which have not. If there are multiple GeneSplicer splice site candidates, the candidate with the highest score is preferred over those with lower scores.
ab initio gene prediction algorithms. The creation and optimization of ab initio gene finders is an active field of study and, as such, many different programs are available to create gene prediction sets. Many of these gene finders have been run on the section of DNA that you will be working on. The results of these analyses are available on the GEP UCSC Genome Browser for your section of DNA. While each program has its strengths and weaknesses, for the purposes of gene model creation (selection of intron/exon boundaries), they should be considered of equal quality. The most common usage of the information created here is a majority rules/vote system. Failing any evidence from basic biology, expression, conservation, or other algorithms, the splice site that was picked most by the different programs would be considered to have the best support.
C. Information for Advanced Annotators
Special situations
There are a few situations that deserve special comments. You may not run across these situations but the comments below can be helpful if you do.
Conservation on one end of an intron. Sometimes when searching for similarity between D. melanogaster exons and your genomic region, you will find one exon with a very good match to the intron/exon junction at one end of the intron and no match at the end of the adjacent exon at the other end of the intron. It is still sometimes possible to find the unaligned site by using a string search instead of a BLAST similarity search. This is probably best explained by example. Consider these two alignments for the first and second exon of some hypothetical gene. In D. melanogaster, the first exon is 68 aa long and the second exon is 28 aa long:
Exon 1 sequence:
1 MDINNEIENIISDIDINIKAQEKLKEQELKAQQYQQNQNK
41 YNPASGPITETQTTTTVVVTKKDSEEET
And alignment to our hypothetical genomic region:
Query 5736 MDINNEIENIISDIDINIKAQEKLKEQELKAQQYQQNQ 5773
MD NN+I NIISDIDINIKAQEKLK+ E ++ + +Q
Sbjct 1 MDFNNQILNIISDIDINIKAQEKLKQNECQSGELDLHQ 38
Exon 2 sequence:
1 SDANVSKTVDLRKIFTPATDAAEILPKN 28
And alignment:
Query 6259 SDSNLSRTVDLRKIFTPATDAPEILPKN 6342
SD+N+S+TVDLRKIFTPATDA EILPKN
Sbjct 1 SDANVSKTVDLRKIFTPATDAAEILPKN 28
Notice the strong alignment at the start of exon 1. This gives good evidence where exon 1 starts, but given its length in D. melanogaster, it may be difficult to find the donor site at the end of exon 1. We would certainly follow the exon length conservation rule above and look downstream about 90 bases (i.e., the last 30 aa or 90 bases of exon 1 are missing from the end of the first alignment), but this just gives the general area where we might expect the end of the exon.
Interestingly, exon 2 starts with a very strong alignment. Thus, when comparing the amino acid (aa) sequence of the D. melanogaster protein with the amino acid encoded in the new species, we have identified two conserved domains separated by a region without conservation. It would be somewhat unlikely that the 5' end of the downstream conserved domain coincides exactly with the 5' end of exon 2. If these two 5' ends do not coincide, we would expect exon 1 should end with at least a few conserved amino acids. Since BLAST did not detect these amino acids (i.e., only the 5' end of exon 1 shows an alignment), it is likely that the number of conserved amino acids at the 3' end of exon 1 is very small (one or two). In these cases, a search for a short DNA sequence that would code for one or two conserved amino acids next to an in-frame splice junction may be fruitful. In the example above, then exon 1 might end with the same 1 or 2 aa as the exon in D. melanogaster.
To start the search we must first find the phase of the acceptor site at the beginning of exon 2. Since there is a very strong alignment that ends at the first amino acid, we expect an acceptor site to be 0, 1, or 2 bases upstream of base 6259. In this example we will assume that the acceptor site at the start of exon 2 immediately precedes the codon for the serine (S). As such, we would look for a DNA sequence to end exon 1 that codes for the amino acid E, and then T (the last 2 amino acids of exon 1) followed immediately by a donor site. If the acceptor site at the beginning of exon 2 had one base between it and then the codon for the serine, we would look for a sequence which has a codon for E, a codon for T, any two bases, and then the donor site. Since the codon table is degenerate, we will need to look for a number of different sequences that could code for ET(donor). Checking the codon table we see that E has two codons, GAG and GAA, and T has four codons, ACT, ACC, ACA, ACG. If these two amino acids were conserved and we want a phase of 0 to match the acceptor of exon 2, then any of 8 different sequences could code for ET(donor):
GAGACTGT GAAACTGT GAGACCGT GAAACCGT GAGACAGT GAAACAGT GAGACGGT GAAACGGT
If the potential region where these amino acids can be is small, it is possible to simply search by eye looking for any of the above sequences. It is also possible to use BLAST to search your sequence if you change some of the parameters to specifically allow for these very short alignments. First, set the word size to a number less than or equal to the length of the sequence you are searching with. Also, be sure to turn OFF the low complexity filter and set a very large Expect threshold (different implementations of BLAST calculate E-values differently when comparing two sequences, it is best to experiment with the version of BLAST you are using to empirically determine the best threshold). Since there are 8 different ways to code for “ET(donor)” you would need to do 8 BLAST searches. (To ensure that the BLAST tool you are using can detect these very small alignments, you may wish to do a positive control search with a sequence you know does exist within your subject to verify that it can be found).
If one of these sequences is found in the correct location (i.e., downstream of the exon 1 alignment but before exon 2 and on the correct strand), and it is in the correct frame (i.e., the same frame as the early exon 1 alignment) and is in the proper phase (i.e., links with the correct frame in exon 2), you have found pretty strong evidence for the end of exon 1 and this site should be picked.
Very small exons. It can be quite difficult to find very small exons. Be sure to increase the Expect threshold until you start to see hits no matter how poor the alignment looks. To avoid sorting through lots of false alignments, you can restrict the region searched. For example, if you have the upstream and downstream exons already mapped, do not search the entire region; rather use the “from:” and “to:” boxes on the BLAST pages to restrict your search to the region between the two mapped exons. You can also try to make the search more sensitive by changing the word size from 3 to 2 but be aware that this may cause you to miss BLAST hits if the size of the sequence you are searching is large so be sure to restrict the area searched if you reduce the word size. Remember, very weak similarity can be the only evidence you will find. However, if it is in the right place and has usable donor and acceptor sites, it is probably identifying the correct exon.
The initial exon is often a very small exon. In these cases, one should not attempt to find the first exon until the second exon has been located. This will help minimize the region where the first exon can be found and give vital evidence to the phase of the donor site. This will then allow you to discount any evidence suggesting that the first exon is outside the region upstream of exon 2 or with an incompatible phase. It is often advisable to use the Small Exons Finder when looking for small exons. In this case, you would look for any candidate exon of the same length as found in D. melanogaster that begins with a methionine codon upstream of a donor site compatible with the exon 2 acceptor. If more than one possible exon is found in the proper region with no evidence to choose among them, pick the one closest to the second exon.
Last and certainly least. It is possible that you may run across situations where you will have ambiguous evidence and must choose between a small number of consistent choices with no evidence to help you decide (this is often the case when using the conservation of exon length rule where there is no expression evidence). In these cases, when all else fails, the policy is to go with the choice that creates the largest protein. The reason for this is that it is better to add a few extra amino acids to a protein than to have a few amino acids missing. This is because, if the amino acids are missing, there is no way to find them in a BLAST search, but BLAST is fairly tolerant of having a few extra amino acids tucked inside an alignment. Thus, it is best to err on the side of extra and not missing amino acids.
It is also possible that you will run across situations where there may be only very weak evidence for one gene model over another yet the weaker model gives a longer protein. To balance these decisions, the GEP has set a policy for the use of the computational donor/acceptor sites when picking your gene model. In general, when picking among a group of consistent intron/exon boundaries where there is no expression or conservation data, choose the longest exon which has a boundary no more than one step (low, medium, high) worse than a boundary that creates a shorter exon. Said another way, when two choices differ by two steps go with the higher valued boundary, and when they differ by one step go for the longer protein. For example, when picking between a donor that makes a longer protein but is unlabeled in the donor/acceptor track versus a donor that makes a shorter protein, only pick the shorter if it is medium or high scoring. On the other hand, if the donor that makes a shorter protein is low quality, pick the boundary that gives the longer protein.
Summary
The following is a list of the important rules for annotation based on the above discussion. All models should follow these rules as much as possible. The rules are listed in the order of importance; the best model will follow a single rule higher on the list at the expense of a single lower rule. However, it is also usually the case that a better gene model will follow many lower rules at the expense of a few higher rules. It is your job as an annotator to find the best gene model that follows as many rules as possible, while giving more credence to higher rules when the evidence is in disagreement, and yet minimizes the total number of discrepancies between the evidence and the final model. For example, you may need to decide between a model that follows many less important rules but breaks a single more important rule and a model that follows a more important rule over the less important rules. This balancing act is where human ability far exceeds computers.
Rules are ranked into four classes:
-
Inviolate rules — rules for which no counter examples have ever been seen or are so extremely rare (say less than 1 in 1,000) that wet bench experimental evidence would be required to convince scientists that this rule should not apply. Unless you are doing wet bench work these rules should never be broken.
-
Important rules — rules for which exceptions are only rarely seen (say less than 1 in 20). You may choose to make a model that does not follow this rule but you must note in your annotation report that this rule was not followed and document all the evidence that led you to not follow this rule.
-
Basic rules — these are rules or observations that are seen more often than not but are also not followed in a significant number of models. You should make models that follow these rules if you can, but be careful not to ignore more important rules just to follow these basic rules. You do not need to document that you did not follow rules of this type.
-
Tie-breaking rules — rules to help make models when all of the more important rules do not help. You may wish to note the use of these rules in your annotation report to help those reviewing your annotations understand why you picked the model you did.
One page summary suitable for printing and keeping with you while you annotate:
Inviolate rules
In Basic Biology
-
CDS of gene must begin with ATG and end with a stop codon, no internal, in-frame stop codons.
-
Exons are found in order along the source DNA.
-
The last two bases of an intron sequence must be AG.
-
Intron sequences should be at least 40 nucleotides (nt).
Important rules
In Basic Biology
-
An intron sequence should begin with GT (GC is the rare exception).
-
GC should be used when use of GT sites breaks important or inviolate rules.
-
-
Use data in RNA-Seq tracks (both mapped reads and splice junction predictions) if available.
In Conservation
-
Conserved amino acids identified by single exon BLAST shown in high quality alignments (i.e., high % identity and properly placed) should be included in exons.
-
The number of exons between informant and new species should be conserved.
-
The organization of exons to generate the various D. melanogaster isoforms should be conserved.
-
Some genes have alternate splice sites for a particular exon that is unique to that isoform. You must find these alternative splice sites and create a gene model for every isoform.
-
Basic rules
In Conservation
-
Identification of conservation should be done in the following order (based on speed, sensitivity, ease of use, and specificity):
-
Protein-DNA blastx or tblastn with increasing Expect thresholds
-
DNA-DNA using the EMBOSS Water tool
-
DNA-DNA blastn using very large E-value cutoff values (e.g., 1010)
-
-
Failing identification of exons by expression or conservation alignments as above, the highly conserved regions identified in the “Comparative Genomics” (multiz) tracks should be checked.
-
Attempt to conserve exon length even if the specific amino acids are not conserved.
-
If it is difficult to identify exons based on the above, repeat the whole process using the gene models generated by the GEP in a close neighbor species as your informant instead of the model found in D. melanogaster.
In Computation
-
Exons that cannot be found by any type of expression or conservation evidence may be identified using predictions from ab initio gene finders
Tie-Breaking rules
In Basic Biology
-
Longer exons are better; include more amino acids in the exons between the start and stop codons.