Cover Sheet
Title
-
Identifying the TSS for a gene in D. eugracilis using sequence alignment with the D. melanogaster ortholog
Objective
-
Identify exon 1 of the Antp gene in D. eugracilis and the region that contains the TSS
-
Use blastn to align nucleotide sequences from D. melanogaster and D. eugracilis
-
Identify the TSS using evidence from blastn
-
Analyze RNA-Seq and TopHat evidence, which may provide additional evidence for the location of a TSS
-
Use a multiple sequence alignment to examine conservation of exon 1 in 28 Drosophila species
-
Use Short Match to identify additional DNA motifs in the core promoter region
Prerequisites
Order
-
Locate the Antp gene in D. eugracilis scaffold
-
Align the exon 1 sequence of Antp-RM from D. melanogaster against the D. eugracilis scaffold
-
Identify the putative TSS for Antp-RM in D. eugracilis
-
Use Short Match to determine if there are canonical DNA motifs in the core promoter region
Resources & Tools
Align the DNA sequence from exon 1 of the Antennapedia gene from D. melanogaster against the D. eugracilis scaffold in order to find the D. eugracilis Transcription Start Site
In Drosophila melanogaster, the gene Antennapedia (Antp) is required for development of the fly’s body plan. It encodes a transcription factor that is part of the Hox gene complex and specifies segment development in the thorax. Absence of Antp expression in the adult fly results in transformation of a leg into an antenna. Ectopic expression in the head results in transformation of an antenna to a leg (or eye to wing).
The Antp gene in D. melanogaster has been annotated, and three TSSs have been defined by FlyBase. We will use information about the TSS for the Antp-RM isoform in D. melanogaster to help us identify the most likely TSS for isoform M of the Antp gene in another Drosophila species, D. eugracilis.
Exercise 1: Locate the Antp gene in D. eugracilis
Let’s begin by locating the Antp gene in D. eugracilis and examining it in greater detail.
-
Open a web browser window and go to the Genomics Education Partnership (GEP) UCSC Genome Browser Mirror and click “Genome Browser” on the left (Figure 1).
-
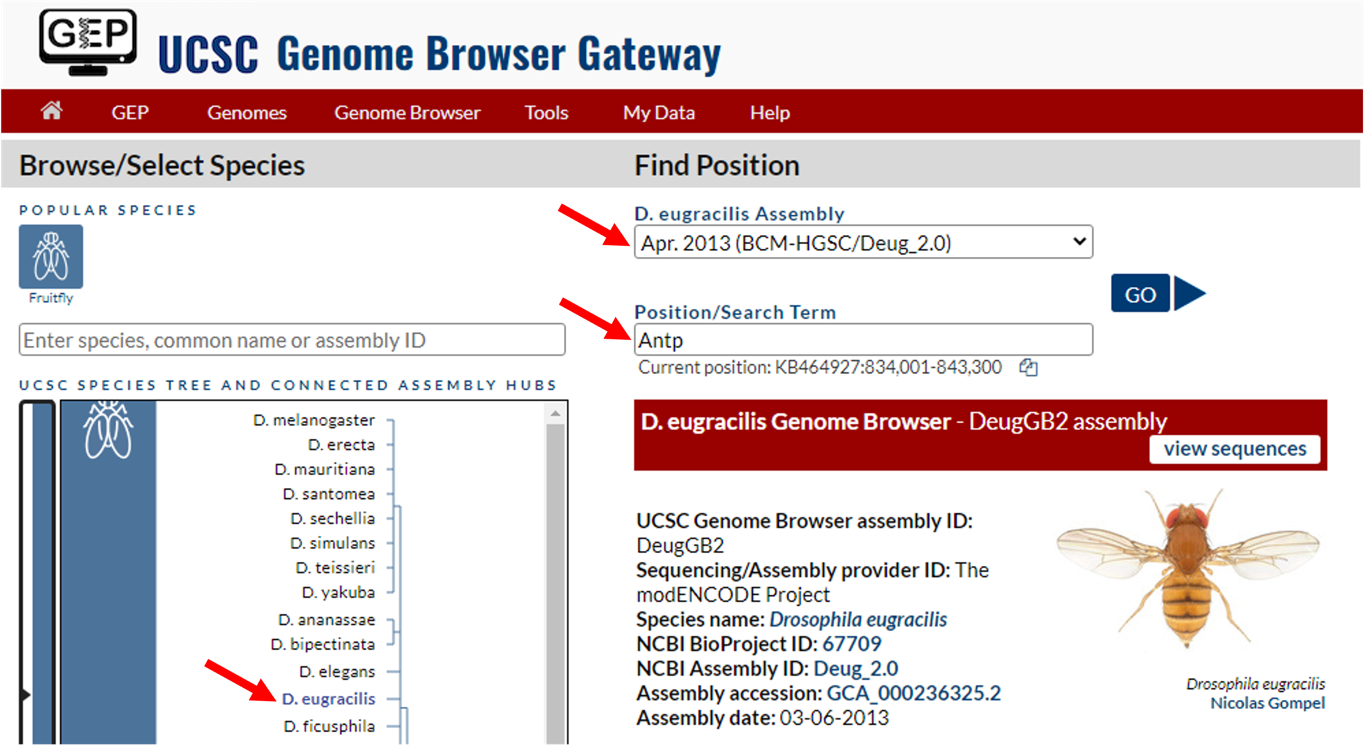
To navigate to the genomic region surrounding the Antp gene in D. eugracilis, select “D. eugracilis” under the “UCSC SPECIES TREE AND CONNECTED ASSEMBLY HUBS” field, select “April 2013 (BCM-HGSC/Deug_2.0)” under the “D. eugracilis Assembly” field, and then enter “Antp” under the “Position/Search Term” field. Click on the “GO” button (Figure 2).
-
Select “Antp-RM at KB465386:38407-150737” from the list “BLAT Alignment of D. melanogaster Transcripts”.
This region on the scaffold KB465386 contains the entire Antp gene, which has 12 isoforms. The suffix “-RM” denotes the name of one isoform that is associated with the gene. Hence Antp-RM corresponds to the M isoform of the Antp gene.
-
We will hide all the evidence tracks and then enable only the subset of tracks that we need. Click on the “hide all” button located below the Genome Browser image. Then, configure the display modes as follows:
-
Under “Mapping and Sequencing Tracks”
-
Base Position: “full”
-
-
Under “Gene and Gene Prediction Tracks”
-
D. mel Transcripts: “pack”
-
-
Under “RNA Seq Tracks”
-
modENCODE RNA-Seq Coverage: “full”
-
modENCODE TopHat Junctions: “full”
-
-
Click on the “refresh” button to update the display
-
-
Let’s zoom in to the region that includes part of the promoter and the beginning of exon 1. Type “KB465386:38,184-38,483” into the “chromosome range, or search terms, see examples” text box, and then click on the “go” button. Note that exon 1 is a non-coding exon. This exon is transcribed into mRNA but does not contain a start codon for translation.
Using RNA-Seq data, which represents the level of mRNA expression, can you identify the TSS for Antp-RM in D. eugracilis?
Using the BLAT Alignment of D. melanogaster Transcripts, RNA-Seq and TopHat evidence, what is the approximate coordinate of the first nucleotide Antp-RM exon 1? For a refresher on RNA-Seq and TopHat, watch the video.
Exercise 2: Use blastn to align exon 1 sequences from D. melanogaster against the D. eugracilis scaffold
You may wonder why we can’t just use RNA-Seq data, TopHat, or promoter motifs such as the Initiator motif to identify the TSS in D. eugracilis. For the Antp gene, these lines of evidence are not sufficient to precisely locate the TSS. Therefore, we will compare DNA sequence from D. eugracilis to the related species D. melanogaster.
Because D. eugracilis and D. melanogaster share a common ancestor, it is likely that their protein-coding genes will show high levels of sequence similarity. The D. melanogaster genome has been fully annotated, so we can obtain evidence for the position of the D. eugracilis TSS by comparing the exon 1 sequence from Antp-RM in D. melanogaster to the D. eugracilis sequence.
To compare the D. melanogaster and D. eugracilis sequences, we will use BLAST (Basic Local Alignment Search Tool).
-
We can use Gene Record Finder to obtain the sequence for exon 1 of Antp-RM in D. melanogaster. Navigate to the Gene Record Finder.
-
Enter “Antp” in the search term box and click on “Find Record”.
-
Scroll down and click on the “Transcript Details” tab. In the “Exon usage map” section, Antp-RM will be highlighted at the top. Note that since we are interested in locating the TSS, we cannot use the polypeptide tab as that only displays the coding (translated) exons used.
-
Scroll down to the exon table and click on the first row (FlyBase ID = 1).
-
A window will open with the sequence for the first transcribed exon of Antp-RM. Copy the sequence for exon 1 marked “>Antp:1”, including the information in the first line.
-
-
Open a new web browser tab and navigate to the NCBI BLAST home page. Click on the “Nucleotide BLAST” image under the “Web BLAST” section. Paste the exon 1 sequence for Antp-RM into the “Enter Query Sequence” text box.
-
Check the box “Align two or more sequences”. The “Enter Subject Sequence” text box will appear.
-
-
To obtain the D. eugracilis sequence for comparison, return to the GEP UCSC Genome Browser tab with the D. eugracilis sequence
-
To ensure that we include the entire scaffold sequence in our search for the TSS, zoom out to view the entire KB465386 scaffold (299,796 bp).
-
-
On the navigation bar at the top of the browser, click on “View”, and select the “DNA” option. Click on the “get DNA” button.
-
Copy the entire sequence — including the information in the first line — and paste into the “Enter Subject Sequence” text box on the blastn page. You should now have the D. melanogaster sequence in the upper box, and the D. eugracilis scaffold sequence in the lower box.
-
-
Change the BLAST algorithm under “Program Selection” to “Somewhat similar sequences (blastn).”
-
Click on the “BLAST” button and wait for the results, which may take about a minute (Figure 3).
-
In the “Sequences producing significant alignments” section, click on the blue text link “DeugGB2_dna range=KB465386:1-299796 5’pad=0 3’pad=0 strand=+ repeatMasking=none” (Figure 3).
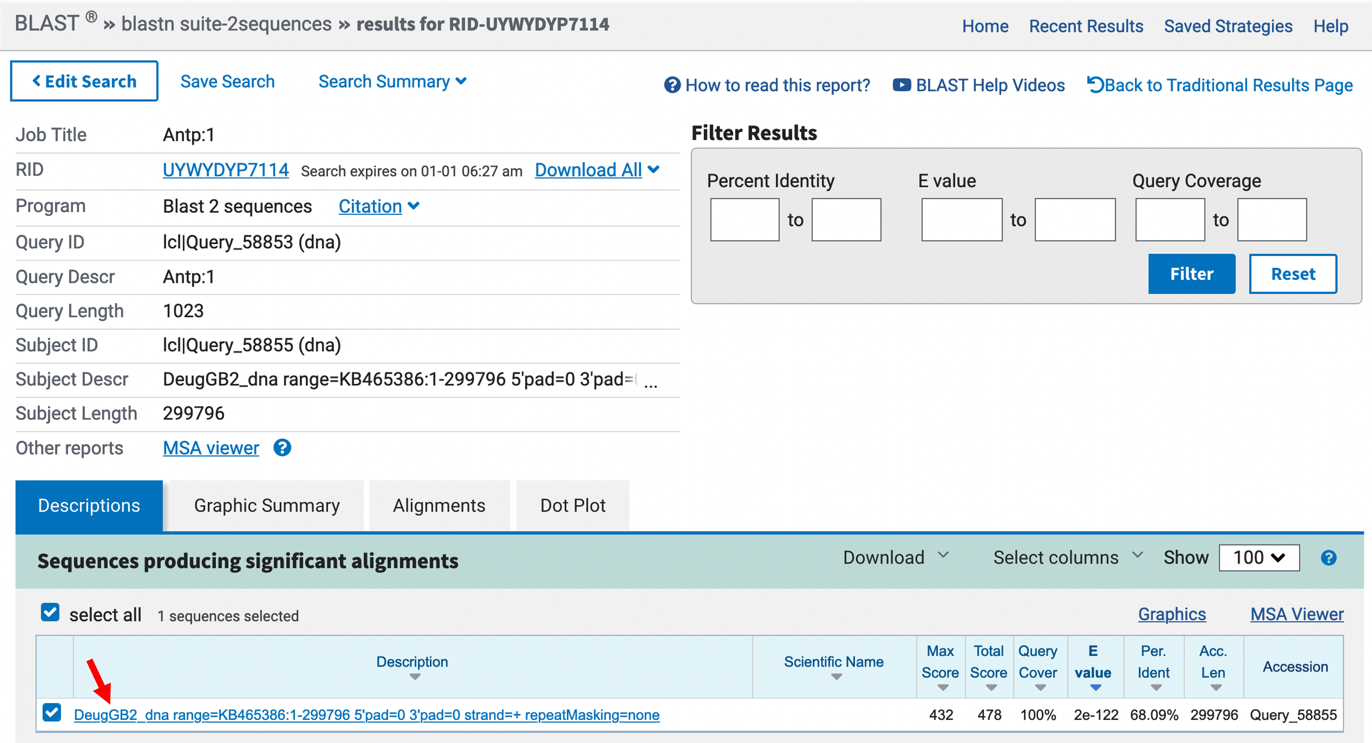
The Expect value (E value) is 2e-122 (Figure 4), which indicates that there less than 1 chance in a googol that the D. melanogaster and D. eugracilis sequences are a random match. Since a googol (10100) is more than the number of protons, electrons, and neutrons in the known universe, we can say with confidence that the sequences are significantly similar.
The sequences are identical for the first 53 nt of Antp-RM exon 1 in D. melanogaster and D. eugracilis, however, there are gaps in the alignment that show that the sequences are not identical for the entire exon.
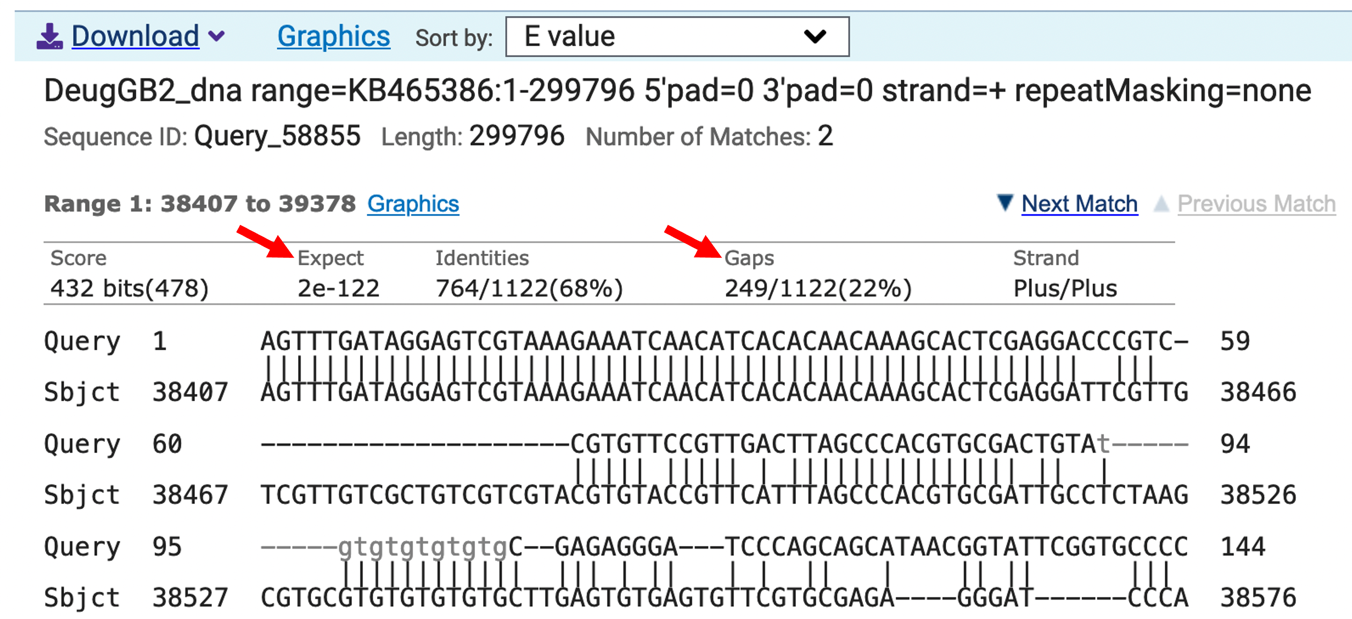
The first nucleotide of exon 1 in D. melanogaster Antp-RM aligns with the D. eugracilis scaffold. Based on the blastn alignment, what is the coordinate of the first nucleotide of exon 1 of Antp-RM in D. eugracilis?
Does the D. eugracilis sequence in the first alignment (“Range 1: 38407 to 39378”) match the D. melanogaster exon 1 sequence exactly?
Exercise 3: Using a multiple sequence alignment to examine exon 1 of the Antp gene in D. melanogaster and D. eugracilis
Wow — it was so easy to find the beginning of exon 1 in D. eugracilis using a blastn alignment. Why did that work so well, and will this approach work with all genes? Let’s look at the D. melanogaster sequence again, using a new tool in the Genome Browser to answer this question.
-
Open a new web browser window, go to the GEP UCSC Genome Browser Mirror, and click “Genome Browser” on the left.
-
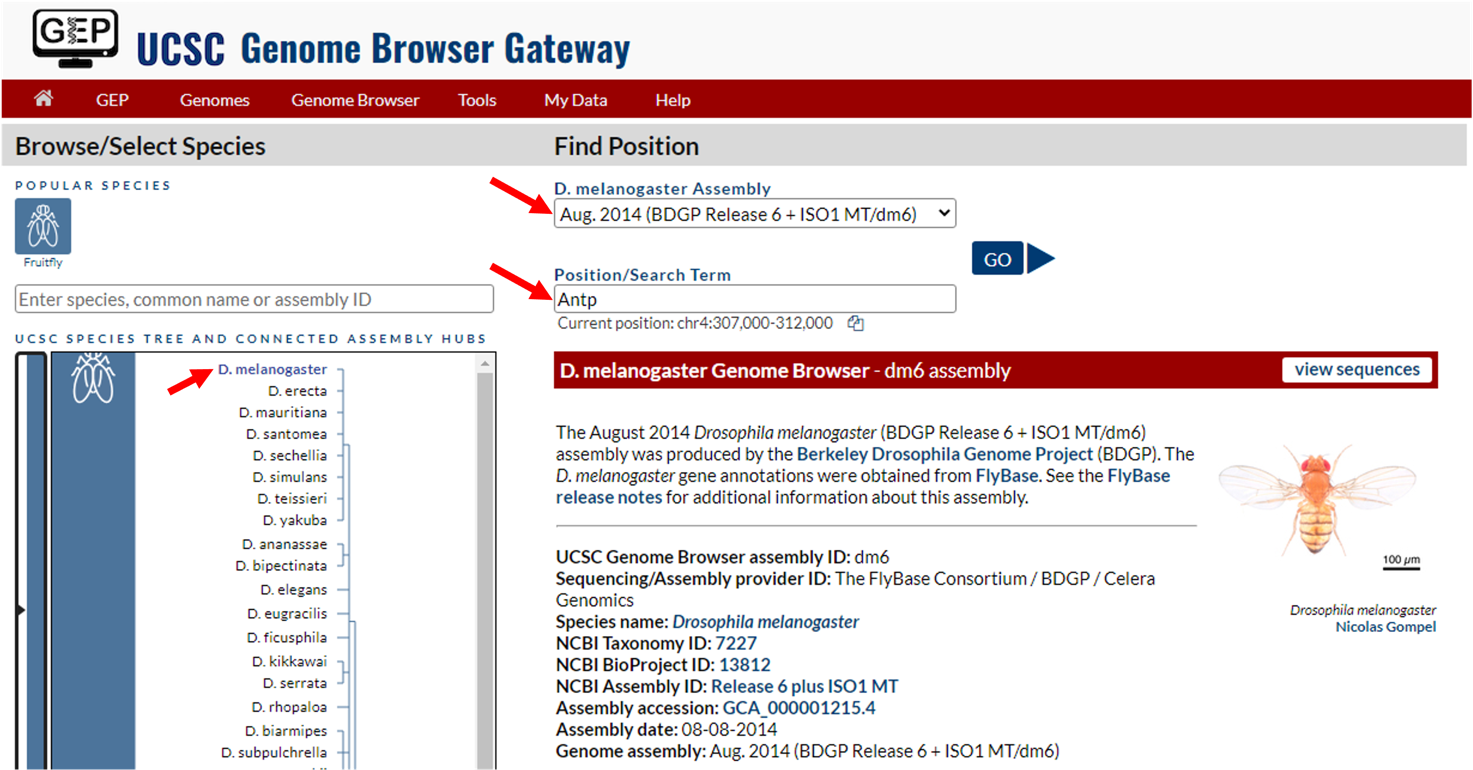
To navigate to the genomic region surrounding the Antp gene in Drosophila melanogaster, select “D. melanogaster” under the “UCSC SPECIES TREE AND CONNECTED ASSEMBLY HUBS” field, select “Aug. 2014 (BDGP Release 6 + ISO1 MT/dm6)” under the “D. melanogaster assembly” field, and then enter “Antp” under the “Position/Search Term” field. Click on the “GO” button (Figure 5).
-
Select “Antp-RM at chr3R:6896253-6999228” from the list of FlyBase Protein-Coding Genes. This region on chromosome 3, denoted “chr3R:6896253-6999228” contains the sequence for Antp-RM. The Antp gene in D. melanogaster also has 12 isoforms.
-
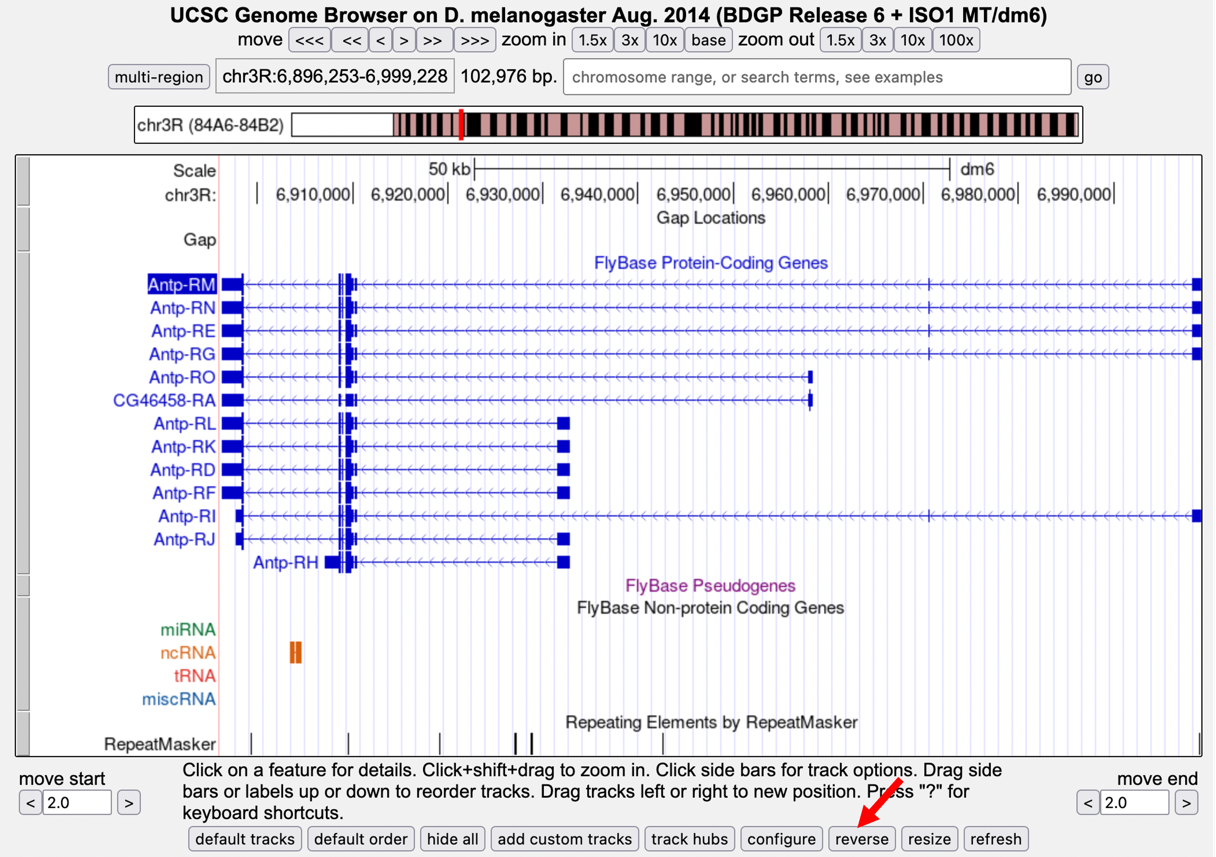
The direction of the arrows in the introns indicates that the Antp gene is on the minus strand. Click on the “reverse” button below the Genome Browser image to reverse complement the chr3R sequence (Figure 6).
-
Click on the “hide all” button located below the Genome Browser image. Then, configure the display modes as follows:
-
Under “Mapping and Sequencing Tracks”
-
Base Position: “full”
-
-
Under “Gene and Gene Prediction Tracks”
-
FlyBase Genes: “pack”
-
-
Under “RNA Seq Tracks”
-
Click on the blue “Combined modENCODE RNA-Seq (Development) (R5)” link
-
Change the “Display mode” to “full”
-
Scroll down to the “List subtracks” section and uncheck the box for “modENCODE RNA-Seq (Plus)”. Then scroll up to the top of the page and click on the “Submit” button to update the display.
-
-
-
Enter “chr3R:6,999,205-6,999,254” into the “chromosome range, or search terms” text box, and then click on the “go” button to zoom in to the beginning of exon 1.
We have now configured the Genome Browser so that we can examine the 5' end of Antp-RM in D. melanogaster. Our goal is to compare this region with the orthologous (evolutionarily related) sequence in D. eugracilis, a “distant cousin” of D. melanogaster.
To do this, we will examine the whole genome multiple sequence alignment of 28 Drosophila species (Figure 7). This evidence track shows the whole genome multiple sequence alignment of 28 Drosophila species constructed by the ROAST program.
-
Configure the Genome Browser as follows:
-
Under “Comparative Genomics”
-
Drosophila Conservation (28 Species): “full”
-
Click on “refresh”
-
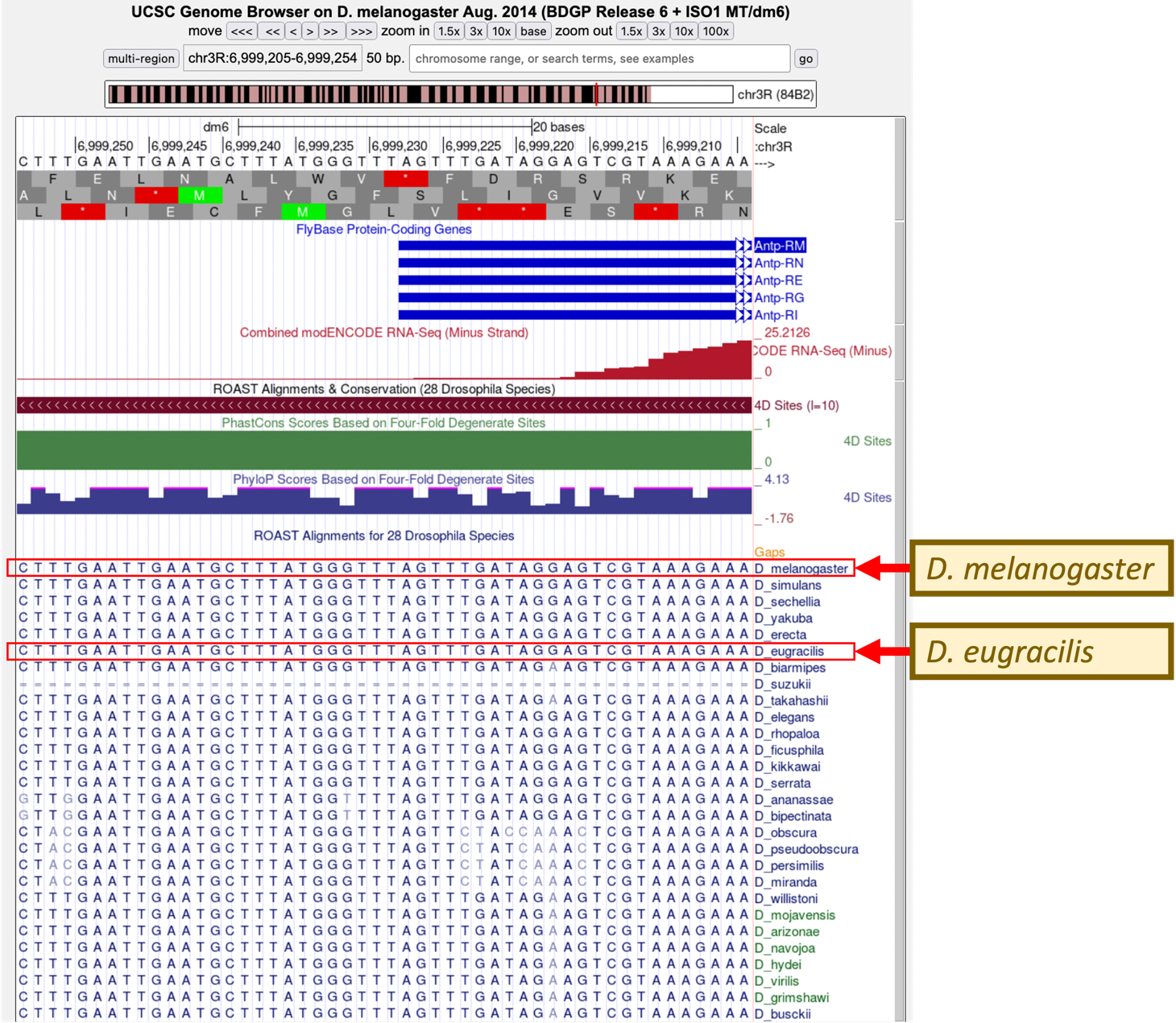
Starting with the “A” nucleotide at the 5' end of exon 1, how many nucleotides can be aligned between D. melanogaster and D. eugracilis?
Under “Comparative Genomics”, click on the blue “Drosophila Conservation (28 Species)” link. Scroll down to the section marked “Display Conventions and Configuration” under “Gap Annotation”. What do the double lines mean in the D. suzukii sequence?
Can we use this strategy to help identify the TSS for any gene?
Using the strategies above we were able to pinpoint the transcription start site of D. eugracilis by comparing its sequence with that of D. melanogaster (Exercise 2). We also investigated the sequence of Exon 1 in multiple other Drosophila species using a comparative genomics track (Exercise 3).
The blastn comparison of the D. melanogaster Antp-RM exon 1 sequence against the D. eugracilis scaffold conducted in Exercise 2 worked well because of the sequence similarity at the beginning of exon 1 in both species. Therefore, using this strategy to identify the first transcribed nucleotide — the transcription start site — will work for any gene in any Drosophila species whose sequence is highly conserved with exon 1 of the orthologous gene in D. melanogaster. When the sequence is not highly conserved between genes, alternative evidence would be necessary for identifying the TSS.
The multiple sequence alignment investigated in Exercise 3 indicates that exon 1 of Antp-RM in D. melanogaster and in D. eugracilis share similar sequences near the TSS. The sequence similarity further supported the identification of the Antp-RM TSS in D. eugracilis.
Let’s take a closer look at the region surrounding the TSS to identify DNA motifs that might play a role in regulating Antp gene expression in vivo.
Transcription factor binding sites: Promoter motifs
Transcription initiation and recruitment of RNA Polymerase II (RNA Pol II) to the promoter region of a gene is achieved with the aid of accessory proteins known as basal transcription factors [Aoyagi et al., 2000]. These basal transcription factors preferentially bind to specific nucleotide sequences within the core promoter region. The nucleotide sequences that are bound by a specific transcription factor can be experimentally determined and used to determine a sequence or DNA motif [Burke and Kadonaga, 1996].
DNA motif is a general term used to describe a short DNA sequence that recurs multiple times throughout the genome and is associated with a particular biological function. Motifs often correspond to binding sites for DNA-binding proteins, and different positions within the motif can show different levels of sequence conservation. Hence, a motif is often represented by a consensus sequence, which captures the variability of the motif instances within a species or among multiple species.
In order to capture the variations that are found in transcription factor binding sites (TFBS), a consensus motif often includes degenerate nucleotide symbols that represent multiple nucleotides. The IUPAC (International Union of Pure and Applied Chemistry) has generated a code to represent degenerate bases [Nomenclature Committee of the International Union of Biochemistry, 1986]. For example, if a site within a sequence motif can be occupied by either of the two pyrimidine bases (C or T), the consensus sequence would be represented by a “Y” at that location. The IUPAC code for degenerate bases is shown in Table 1.
| Symbol | Description | Nucleotides |
|---|---|---|
R |
Purine |
A or G |
Y |
Pyrimidine |
C or T |
W |
Weak |
A or T |
S |
Strong |
C or G |
M |
Amino |
A or C |
K |
Keto |
G or T |
H |
Not G |
A, C, or T |
B |
Not A |
C, G, or T |
V |
Not T |
A, C, or G |
D |
Not C |
A, G, or T |
N |
Any |
A, C, G, or T |
It is not unusual for sequence motifs to possess sites in which two or more bases appear at similar frequencies (e.g., K and Y in the TCAKTY motif in Table 2). In fact, these differences in the motif instances could help modulate gene expression [Spivakov et al., 2012].
| Motif | Consensus | Position Relative to TSS |
|---|---|---|
BREu |
SSRCGCC |
-38 |
TATA Box |
TATAWAAR |
-31 or -30 |
BREd |
RTDKKKK |
-23 |
Inr |
TCAKTY |
-2 |
MTE |
CSARCSSAAC |
+18 |
DPE |
RGWYVT |
+28 |
Ohler_motif1 |
YGGTCACACTR |
NA |
DRE |
WATCGATW |
NA |
Ohler_motif5 |
AWCAGCTGWT |
NA |
Ohler_motif6 |
KTYRGTATWTTT |
NA |
Ohler_motif7 |
KNNCAKCNCTRNY |
NA |
Ohler_motif8 |
MKSYGGCARCGSYSS |
NA |
The Inr motif is sometimes found at the beginning of exon 1 of genes in Drosophila. At which two nucleotide positions are there degenerate bases in the Inr motif consensus sequence?
The most common core promoter motifs for Drosophila are shown in Table 2. For each motif, the consensus sequence is shown as well as the position of the motif relative to the TSS if they are known. Motifs that are enriched in broad promoters (e.g., the Ohler motifs 1, 5, 6, 7, and 8) do not have a known position relative to the TSS because broad promoters contain multiple TSSs [Ohler et al., 2002]. Note that the first transcribed nucleotide is assigned the position of +1.
A core promoter region can contain zero or more core promoter motifs. Some core promoter motifs are associated with the same basal transcription factor. For example, the Inr, TATA box, MTE, and DPE motifs are all associated with Transcription Factor II D (TFIID) [Theisen et al., 2010], and the BREu and BREd motifs are associated with the Transcription Factor II B (TFIIB) [Deng and Roberts, 2006]. The combination of motifs in a given promoter region can be a critical determinant in regulating gene expression [Juven-Gershon et al., 2006].
It is important to realize that promoters can vary widely, possessing different combinations of motifs [Vo ngoc et al., 2017]. For example, different isoforms of a single gene can have different promoters, each with a different combination of motifs. Screening of promoters for core promoter motifs using computational tools has shown that many promoters do not have any of the known core promoter motifs shown in Table 2 [Sloutskin et al., 2015]. Nevertheless, it is important to probe a putative TSS site location for the presence of these motifs as it provides more supporting evidence for the TSS annotation.
Exercise 4: using the short match feature to search for motif sequences
-
Go back to the GEP UCSC Genome Browser tab to examine the D. eugracilis sequence further. You will need to click back a couple of times to get to the browser from the DNA sequence view.
-
Enter “KB465386:38,352-38,451” into the “chromosome range, or search terms” text box, and then click on the “go” button to navigate to the region surrounding the TSS of Antp.
-
Ensure that your browser is configured as follows:
-
Base Position: “full”
-
D. mel Transcripts: “pack”
-
modENCODE RNA-Seq Coverage: “full”
-
Short Match: “pack”
-
-
Under “Mapping and Sequencing Tracks”, click on the blue “Short Match” link
-
Enter the Inr consensus sequence “TCAKTY” in the “Short (2-30 base) sequence” text box
-
Click “Submit”
-
Is there an Inr motif in this region? If so, what are the coordinates for the motif? Remember that motifs on the minus strand will have coordinates preceded by a minus sign (-), and motifs on the plus strand will have coordinates preceded by a plus sign (+).
-
Under “Mapping and Sequencing Tracks”, click on the blue “Short Match” link
-
Enter the BREd consensus sequence “RTDKKKK” in the “Short (2-30 base) sequence” text box
-
Click on “Submit”
-
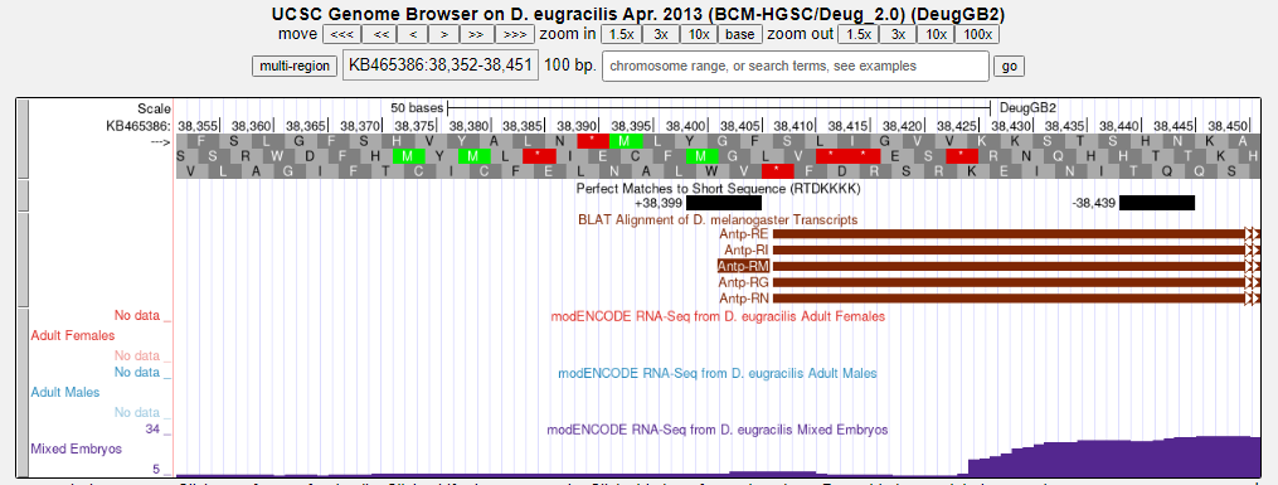
The “Short Match” track shows there are two matches to the RTDKKKK sequence — one on the plus strand and one on the minus strand (Figure 8). Note that we expect the core promoter motifs to be in the same orientation as the direction of transcription. In addition, given the information in Table 2, we can see that BREd motifs are usually found at position -23 relative to the TSS in Drosophila. Let’s investigate this region further.
Analyze the BREd motif instances on the Short Match track (you may need to zoom in). Is there a BREd motif at -23 relative to the start of transcription (+1)? Note that the core promoter motif should be in the same orientation as the direction of transcription. Based on the information about the characteristics of core promoter motifs above, is there a canonical BREd motif for the core promoter of Antp-RM?
Perform Short Match searches for the rest of the core promoter motifs in Table 2. Are any of these motifs located in the correct positions relative to the TSS of Antp-RM?
Wrapping up
Although promoter motifs are commonly found in the promoter regions of protein-coding genes, there are many genes that do not appear to use such motifs for initiating transcription. Finding motifs within the predicted promoter region of a gene is exciting and confirms our current understanding of transcription initiation and regulation. The lack of promoter motifs for some genes, however, tells us that we still have a lot to learn about how the cell turns gene expression on and off!
Bibliography
-
[Aoyagi et al., 2000] Aoyagi, N. & Wassarman, D. A. Genes Encoding Drosophila melanogaster RNA Polymerase II General Transcription Factors: Diversity in TFIIA and TFIID Components Contributes to Gene-specific Transcriptional Regulation. J. Cell Biol. 150, 45–50 (2000).
-
[Burke and Kadonaga, 1996] Burke, T. W. & Kadonaga, J. T. Drosophila TFIID binds to a conserved downstream promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10, 711–724 (1996).
-
[Nomenclature Committee of the International Union of Biochemistry, 1986] Nomenclature Committee of the International Union of Biochemistry. Nomenclature for incompletely specified bases in nucleic acid sequences. Proc. Natl. Acad. Sci. U. S. A. 83, 4–8 (1986).
-
[Spivakov et al., 2012] Spivakov, M. et al. Analysis of variation at transcription factor binding sites in Drosophila and humans. Genome Biol. 13, R49 (2012).
-
[Ohler et al., 2002] Ohler, U., Liao, G., Niemann, H. & Rubin, G. M. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3, research0087.1 (2002).
-
[Theisen et al., 2010] Theisen, J. W. M., Lim, C. Y. & Kadonaga, J. T. Three Key Subregions Contribute to the Function of the Downstream RNA Polymerase II Core Promoter. Mol. Cell. Biol. 30, 3471–3479 (2010).
-
[Deng and Roberts, 2006] Deng, W. & Roberts, S. G. E. Core promoter elements recognized by transcription factor IIB. Biochem. Soc. Trans. 34, 1051–1053 (2006).
-
[Juven-Gershon et al., 2006] Juven-Gershon, T., Cheng, S. & Kadonaga, J. T. Rational design of a super core promoter that enhances gene expression. Nat. Methods 3, 317–322 (2006).
-
[Vo ngoc et al., 2017] Vo Ngoc, L., Wang, Y., Kassavetis, G. A. & Kadonaga, J. T. The punctilious RNA polymerase II core promoter. Genes Dev. 31, 1289–1301 (2017).
-
[Sloutskin et al., 2015] Sloutskin, A. et al. ElemeNT: a computational tool for detecting core promoter elements. Transcription 6, 41–50 (2015).